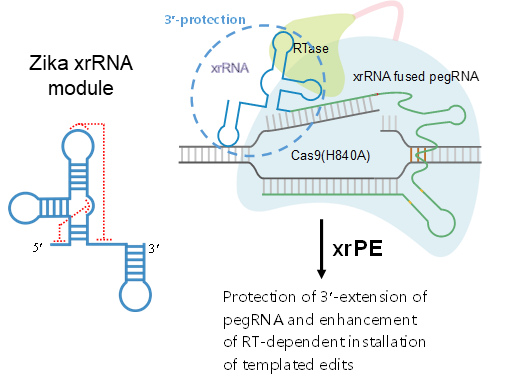
the prime editors (pes) have shown great promise for precise genome modification. however, their suboptimal efficiencies present a significant technical challenge. by appending a viral exoribonuclease-resistant rna motif (xrrna) to the 3-extended portion of pegrnas for their increased resistance against degradation, zhang et al. develop an upgraded pe platform (xrpe) with substantially enhanced editing efficiencies in multiple cell lines. a pan-target average enhancement of up to 3.1-, 4.5- and 2.5-fold in given cell types is observed for base conversions, small deletions, and small insertions, respectively. additionally, xrpe exhibits comparable edit:indel ratios and similarly minimal off-target editing as the canonical pe3. of note, parallel comparison of xrpe to the most recently developed epegrna-based pe system shows their largely equivalent editing performances. this study establishes a highly adaptable platform of improved pe that shall have broad implications.
dr. jianghuai liu from model animal research center at the medical school of nanjing university, dr. xingxu huang from shanghaitech university, and dr. xiaolong wang from northwest a&m university, together with a number of collaborators from several institutions, contributed to this project. the work has been published in nature communications (“enhancement of prime editing via xrrna motif-joined pegrna,” 2022).



